Weak lensing pipeline guide¶
This is a brief introduction on how to use this package in pipeline mode. Please note that this Tutorial describes a simple scenario, in case you encounter problems or unexpected behaviour, inspect the source code, or contact us directly.
All of these scripts have a set of dedicated runtime flags, e.g. to skip processing random points.
Measuring the weak lensing data vector¶
Some pre-defined scripts are located in bin/redpipe/
Define the parameters and inputs as described in Config files explained
Exectute
mkbins.py, there are some flags available, e.g. in case you don’t have any random points, you can use the--norandsflag to skip them.This script loads the input files, and splits them into parameter bins and JK-patches, and writes them to disk in a format which is understood by XSHEAR
The input files are written to
[custom_data_path]/xshear_in/[tag]/Run XSHEAR on the created input files. Depending on the choice of source galaxy catalog use either
xshear.pyfor normal runs, andxshear_metacal.pyfor METACALIBRATION.Note that this step might take a very long time, consider running it on a dedicated computing cluster
This step support OpenMP style parralelization to assign the calculation of separate K-means regions to multiple cores. As a backup solution, it also supports splitting it up to multiple individual tasks via the flags
--nchunk(number of chunks), and--ichunk(ID of chunks).The output files are written to
[custom_data_path]/xshear_out/[tag]/Extract the lensing profile from the xshear results via
postprocess.pyBy default the extracted quantity is
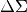 , but
, but 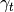 can also be
extracted by re-defining attributes of
can also be
extracted by re-defining attributes of StackedProfileContainer.The results are written to
[custom_data_path]/results/[tag]/The resulting lensing profiles are written as
_profile.dat, and the corresponding Jackknife covariance is saved as_cov.dat.In case random points are also defined, there are three types of output files: lens, randoms and subtracted.
Boost factor estimates from P(Z) decomposition¶
Some pre-defined scripts are located in bin/tools/photoz/, but note that some of the
later steps can be controlled when run in an interactive session.
Extract
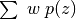 and
and 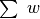 for each K-means region via
for each K-means region via
extract_full_pwsum.py.Note that this step might take a very long time, consider running it on a dedicated computing cluster
This step support OpenMP style parralelization to assign the calculation of separate K-means regions to multiple cores. As a backup solution, it also supports splitting it up to multiple individual tasks via the flags
--nchunk(number of chunks), and--ichunk(ID of chunks).Furthermore the
--ibinflag restricts the calculation to a single parameter bin.The output files are written to
[custom_data_path]/xshear_out/[tag]/as npz filesCombine K-means regions into a
PDFContainerwith Jackknife regions usingextract_full_PDF.py(note that this is a difference between one-patch and all-except-one-patch).Perform the P(z) decomposition as outlined in
mkboost.py.Note that this step in practice requires to set the parameter bounds for the fit, and for this reason it’s best run in an interactive mode. The script is only intended to serve as an example on how the decomposition can be performed.