API Reference¶
DES Y3 wrappers¶
SOMPZ¶
Combine shear profiles and apply calibrations¶
WEIGHTS must be from the base input dataset for Random points!!! |
Input file manipulation¶
The bulk of the action is performed by the main writer class:
|
XSHEAR style input file creator |
Additional helper functions are defined below.
Specify observational fields for the lens catalog:
Return lists of input files |
|
|
Applies RA, DEC cut based on DES field boundaries |
|
Extracts field boundaries from project |
Define K-means and Jackknife regions on the sphere:
Defines 2D patches on the sky via spherical k-means |
|
Assigns a Jacknife (JK) label to the points based on the passed centers |
|
Extracts JK-labels from k-means label array |
Load and prepare lens and random point catalogs:
|
Loads lens catalog from fits file |
Loads lens data and defines sub-selections for different parameter bins |
|
|
Loads random point catalog from fits file |
Loads random points and defines sub-selections for different parameter bins |
XSHEAR wrapper¶
xshear config file writer¶
the main writer functions:
|
Writes simple XSHEAR config file based on |
Writes custom XSHEAR config file |
addittional helper functions:
Load settings for unsheared METACAL run with pairlogging |
|
Load settings for unsheared METACAL run with out pairlogging |
|
|
appends lines to config file |
|
Returns the source-lens pair logging config part of an XSHEAR config file |
|
Returns the radial binning config part of an XSHEAR config file |
|
Returns the shear config part of an XSHEAR config file |
|
Returns the cosmology config part of an XSHEAR config file |
Calculates METACAL-style sheared file names |
Running xshear¶
|
Creates configuration dictionary which can be passed to multiprocessing map_async() |
|
Calls xshear in a single process |
Executes serial calculation for each chunk (simple for loop) |
|
|
OpenMP style parallelization for xshear tasks |
Random rotations of the source catalog¶
|
runs one single rotation of the source catalog with METACAL SELECTION RESPONSES |
|
performs the random rotations serially and saves them to file |
Performs the random rotations and saves them to file |
The catalog rotator object
|
Loads shear catalog, and saves a randomly rotated version |
additional functions:
|
Radnom generates seeds for random rotations using the master seed |
|
2D counterclockwise roation matrix |
Postprocessing XSHEAR output¶
High level wrapper for postprocessing single parameter bins:
Extracts StackedProfileContainer from xshear output |
Which wraps the main container class, responsible for most of the postprocessing:
Object Oriented interface for stacked shear and |
Some other useful functions¶
Extract area-weighted radial bins centers for the lensing measurement:
|
Calculates nominal edges and centers for logarithmic radial bins(base10 logarithm) |
Jackknife covariance between different parameter bins:
Calculates the Covariance between a list of profiles |
XSHEAR results I/O¶
The main reader function
|
Reader for xshear output if style is set as both |
Addtitional helpers for I/O:
Reads and interprets xshear output from a single file |
|
Reads and interprets xshear output from many smaller files |
|
Processes many smaller xshear output files via xread |
Reads xshear output from file |
|
Reads xshear output from multiple files, and concatenates them |
|
reads raw xshear output from metacal sheared runs |
|
reads raw xshear output from metacal sheared runs |
|
Write an xshear style lens catalog to file |
Reads postions based on the list of filenames passed |
Cluster member contamination estimates¶
Tool to package information about what to do:
Calculate average photo-z P(z) PDF:¶
Additional useful tools:
P(z) and Boost container object¶
The Main Container Object is:
The JK-region collation is performed by:
Classes for the P(z) decomposition:
The JK-region collation is performed by:
Additional useful tools:
Useful tools¶
tools.catalogs¶
|
Converts potentially nested record array (such as a FITS Table) into Pandas DataFrame |
additional functions:
|
Assigns the dtypes to the flattened array |
|
Copies the record array into a new recarray which has only 1-D columns |
tools.selector¶
Manipulate arrays and parameter distributions
|
Applies selection to array based on the passed parameter limits |
|
Matches two D-dimensional distributions by reweighting individual objects |
|
Divides a list into N roughly equal chunks |
|
Calculates x / y for arrays, setting result to zero if x ~ 0 OR y ~ 0 |
# TODO Visualization
example function¶
This is a one line description |
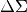 calculated via xshear
calculated via xshear