xpipe.xhandle.shearops.StackedProfileContainer¶
- class xpipe.xhandle.shearops.StackedProfileContainer(info, data, labels, ncen, lcname=None, metadata=None, metatags=None, Rs=None, weights=None, shear=False, **kwargs)[source]¶
Object Oriented interface for stacked shear and
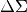 calculated via xshear
calculated via xshearFeatures
Jackknife based covariance estimation
Calculate shear, DeltaSigma, and other averaged quantities
Simple arithmetic with shear profiles:
+, -, *, /Integration with Metacalibration style selection responses
Overview
The lensing profile is calculated in two stages:
First, for each Jackknife-region defined by the
labels(that is for all-except-one region) the lensing profile is calculated, and stored inself.*_sub(These are the JK-subprofiles).Using these JK-subprofiles, the mean lensing profile and the corresponding covariance is calculated via blockwise omitt-one Jackknife resampling.
In practice both of these steps are performed when calling self.prof_maker(), and the results are stored in the below varaiables. The default quantity calculated is the DeltaSigma:
self.rr: radial centers self.dst: E-mode lensing signal self.dst_err: error for E-mode lensing signal self.dst_cov: covariance for E-mode lensing signal self.dst: B-mode lensing signal self.dst_err: error for B-mode lensing signal self.dst_cov: covariance for B-mode lensing signal
A benefit of this two-step method, is that it allows for the quick calculation of errors on the differences, ratios and other transformations of shear profiles.
In particular, the self.composite function allows four basic operations
+, -, *, /between shear profiles, while the self.multiply function allows\*, /with integers or 1-D arrays (in which case each element is a value for a lens)E.g to calculate
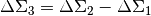 ,
we only need to compute the differences of JK-subprofiles and
then re-compute the JK-mean, and JK-covariance.
,
we only need to compute the differences of JK-subprofiles and
then re-compute the JK-mean, and JK-covariance.In practice the above operation is achieved by:
# the operation is performed in-place, hence the deepcopy prof3 = copy.deepcopy(prof2) prof3.composite(prof1, operation="-")
Changing the physical quantity computed by self.prof_maker() is possibly by modifying the columns used by the stacking algorithm. The default values used for DeltaSigma are:
# default values for stacked DeltaSigma self.dst_nom = 4 self.dsx_nom = 5 self.dst_denom = 6 self.dsx_denom = 7 self.e1_nom = 10 self.e2_nom = 11 self.meta_denom = 3 self.meta_prefac = 2
To calculate shear one would need to change the above to:
self.dst_denom = 8 self.dsx_denom = 9 self.meta_denom = 3 self.meta_prefac = 3
Further information on the column positions are found in the xread documentation…
- Parameters
info (np.ndarray) –
infotable from xreaddata (np.ndarray) –
datatable from xreadlabels (np.array) – JK-region labels
ncen (int) – number of JK regions in total
lcname (str) – tag string
metadata (list of np.ndarray) – sheared versions of
datametatags (list of str) – sheared tags
Rs (float) – metacal selection response (constant) which will be used instead of metadata / metatags
- composite(other, operation='-', keep_rr=True)[source]¶
Calculate the JK estimate on the operation applied to the two profiles
Possible Operations:
“-”: self - other
“+”: self + other
“*”: self * other
“/”: self / other
The results is updated to self. Use deepcopy to obtain copies of the object for storing the previous state.
- Parameters
other (StackedProfileContainer) – an other profile…
operation (str) – string specifying what to do…
keep_rr (bool) – whether to reset radial values to placeholder
- drop_data()[source]¶
resets the
info,dataandlabelscontainers toNoneThis reduces the size of the object in memory, but cannot re-stack the lenses anymore. Leaves the profile, JK-subprofiles and ancillary quantities unaffected
- classmethod from_sub_dict(sub_dict)[source]¶
Reconstructs the object from a dictionary
The resulting object does not contain the base data, only the profile, JK-subprofiles and ancillary quantities
- multiply(val, keep_rr=True)[source]¶
Calculate the JK estimate on profile times the specified “val”,
Operation performed in-place. Use deepcopy to obtain copies of the object for storing the previous state.
- Parameters
val (float or np.array) – value to multiply by
- prof_maker(weights=None)[source]¶
Calculates the Jackknife estimate of on stacked profile and on the corresponding covariance
- Parameters
weights (np.array) – weight for each entry in the lens catalog